Research
Our group seeks to understand the origin and principle of life by constructing artificial molecular systems through Darwinian evolution and design.
Over the past few decades, molecular biology has revealed most of the sophisticated molecular systems in the cell. However, researchers have not yet succeeded in constructing cells from scratch, nor do they know how life emerges from a collection of molecules. Our ultimate goal is to construct artificial molecular systems that are indistinguishable from living cells and to understand what life is.
To that end, we are now tackling these three projects.
- Darwinian evolution of RNA-protein self-replication system
- Designing and constructing self-regenerative molecular systems
- Applications using the constructed systems
Darwinian evolution of RNA-protein self-replication system
To understand how unique features of biology (e.g., diversity, complexity, etc.) arise from a collection of molecules, we have constructed an evolvable RNA-protein replication system and conducted long-term evolutionary experiments. The system consists of a reconstituted E. coli translation system (PURE system) and a single-stranded RNA (artificial RNA genome) encoding RNA replicase. The whole system is encapsulated in a micro-scale water-in-oil emulsion or liposomes. When we repeated replication by periodically adding droplets containing nutrients (the translation system) into the emulsion system (Fig. 1-1, Ichihashi et al. 2013), the RNA continuously evolved by accumulating mutations introduced by replication errors.
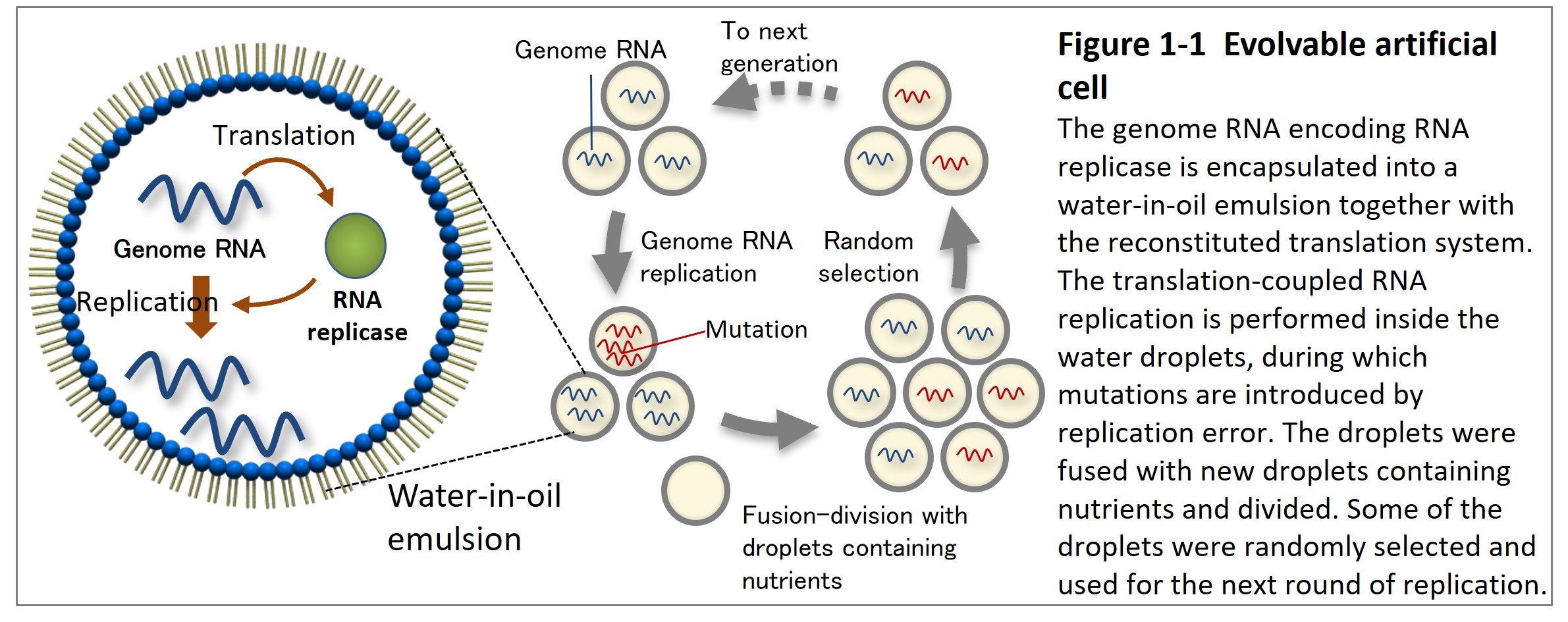
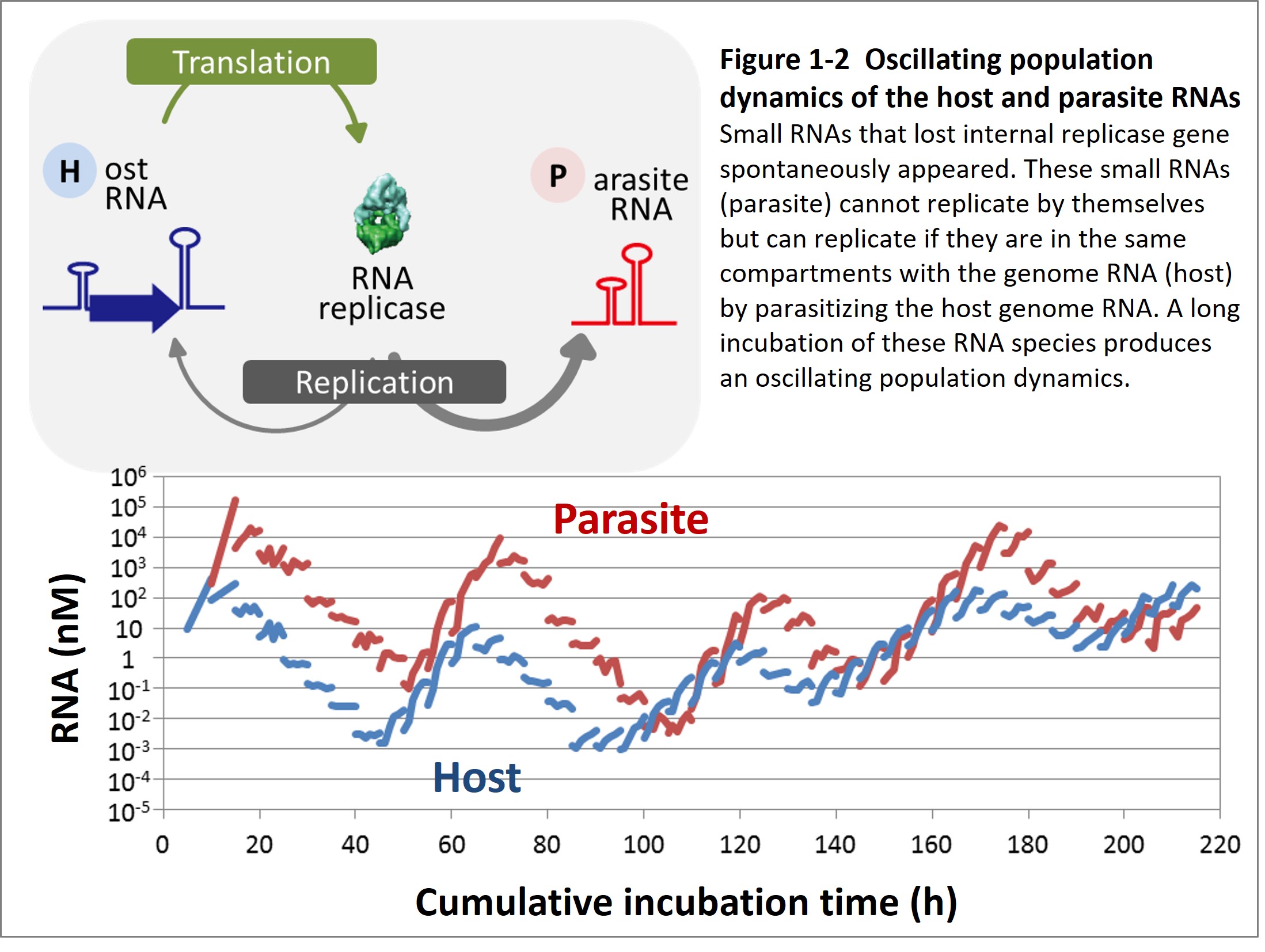
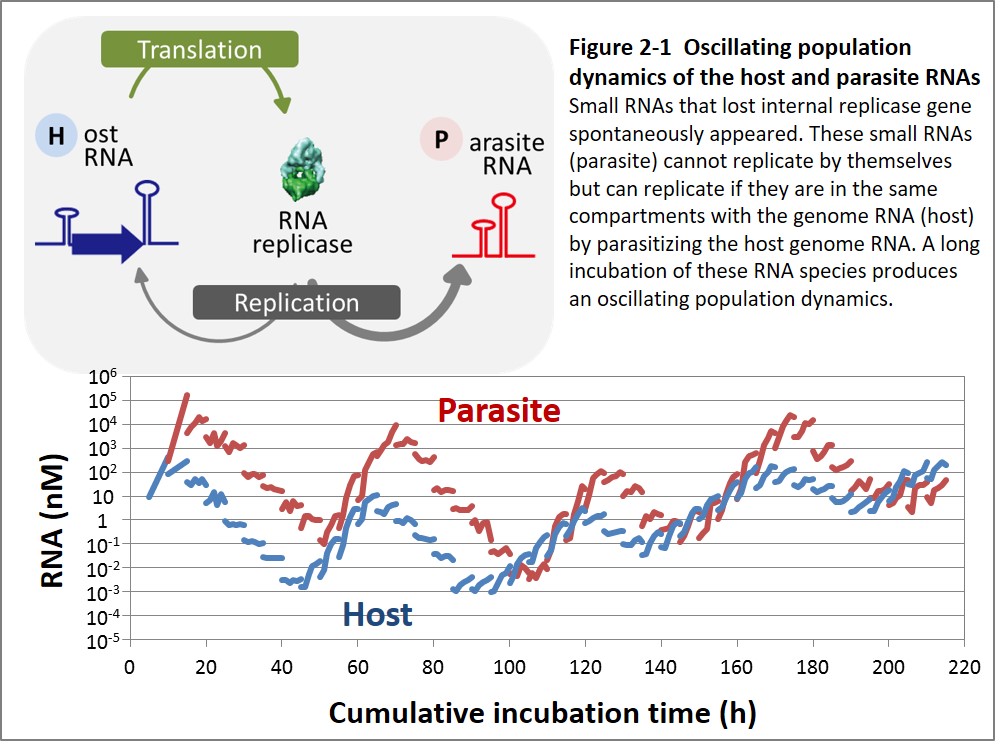
To date, the experimental evolution of the RNA-protein replication system has revealed some evolutionary phenomena that were thought to be unique to living things. First, we found that parasitic RNA species that lost the replicase gene appeared during evolution and generated oscillating population dynamics sometimes observed in nature between host and parasite or prey and predator (Figure 1-2, Bansho et al. 2016).
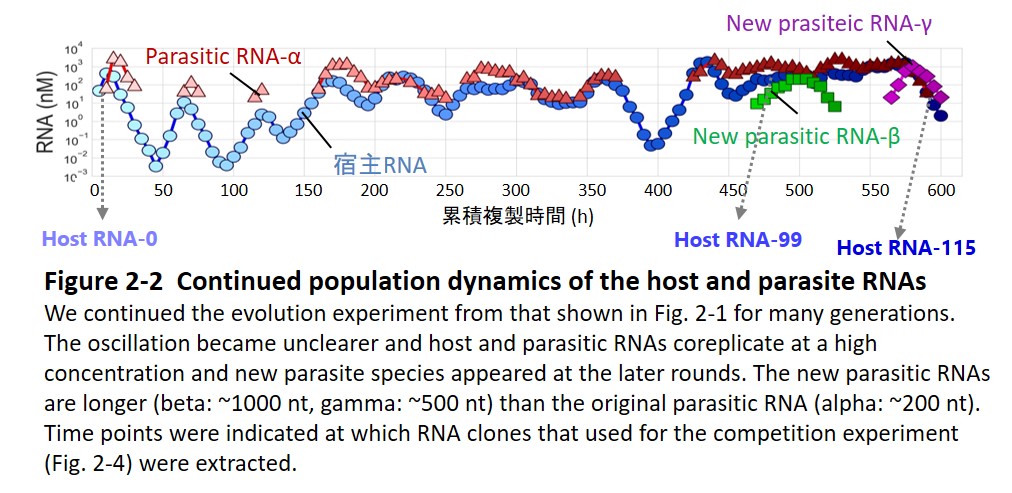
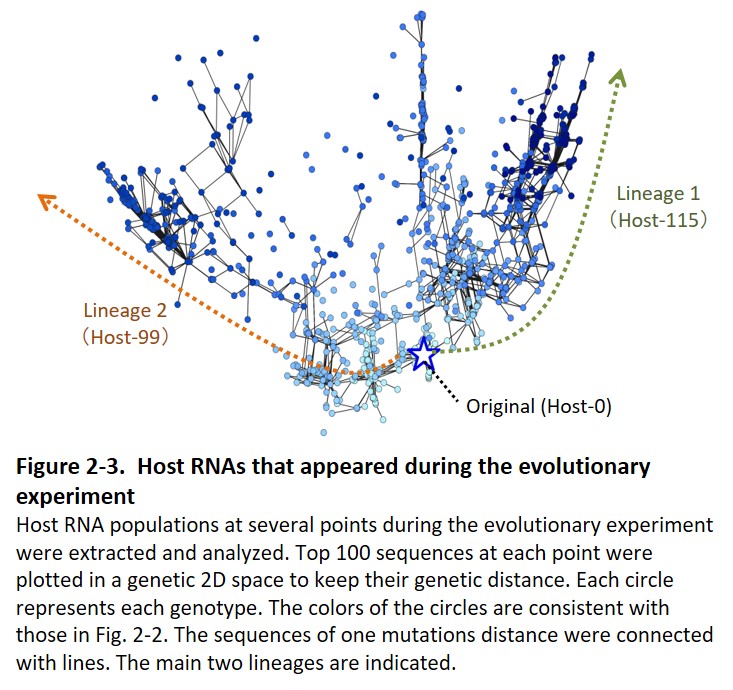
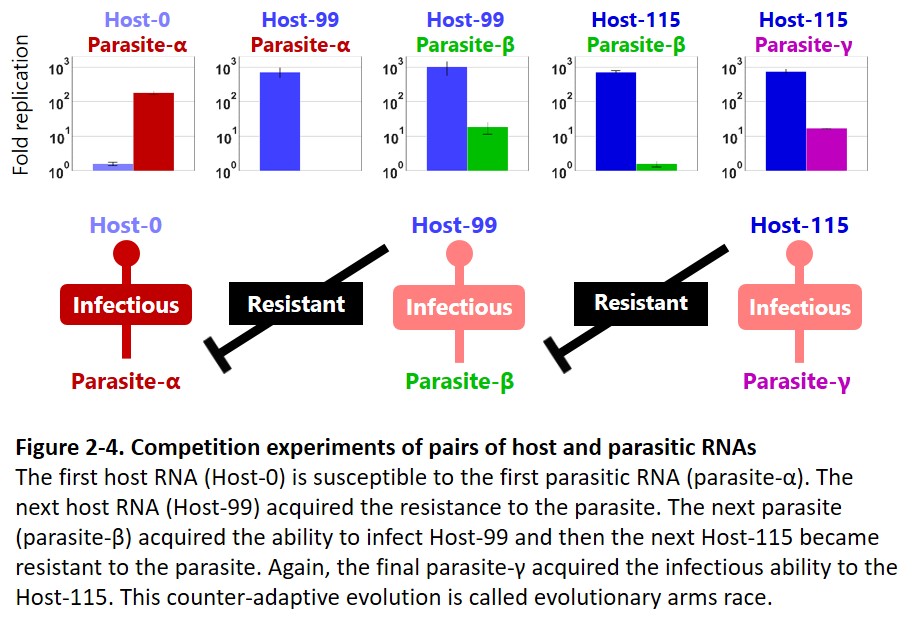
Second, the host and parasitic RNA started an evolutionary arms race with the host genomic RNA (Furubayashi et al. 2020, Kamiura et al. 2022) and diversified into up to five distinct lineages (Fig. 1-3). The differentiated RNA species form an interdependent replication network in the final population (Fig. 1-4, Mizuuchi et al. 2022).
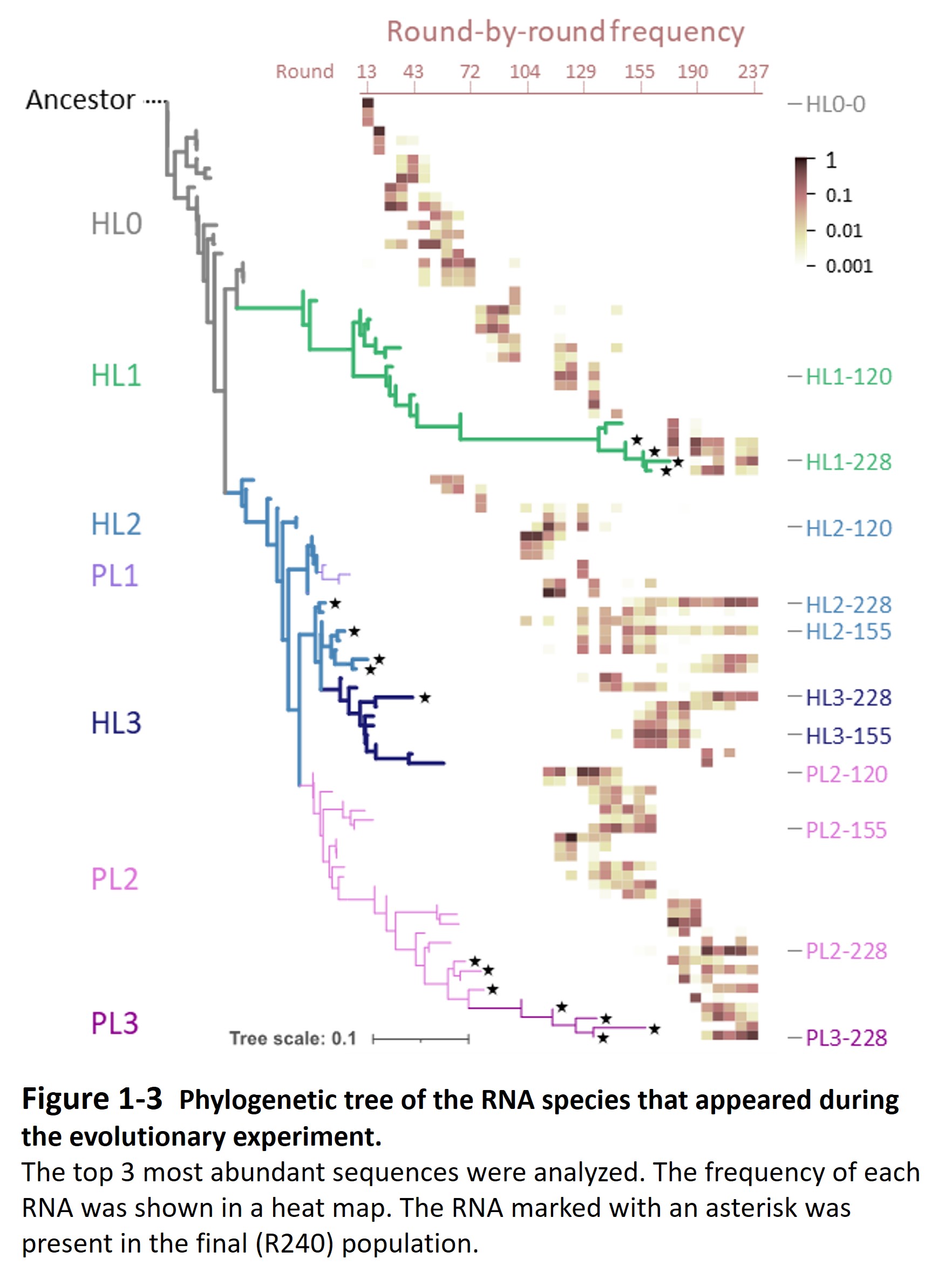
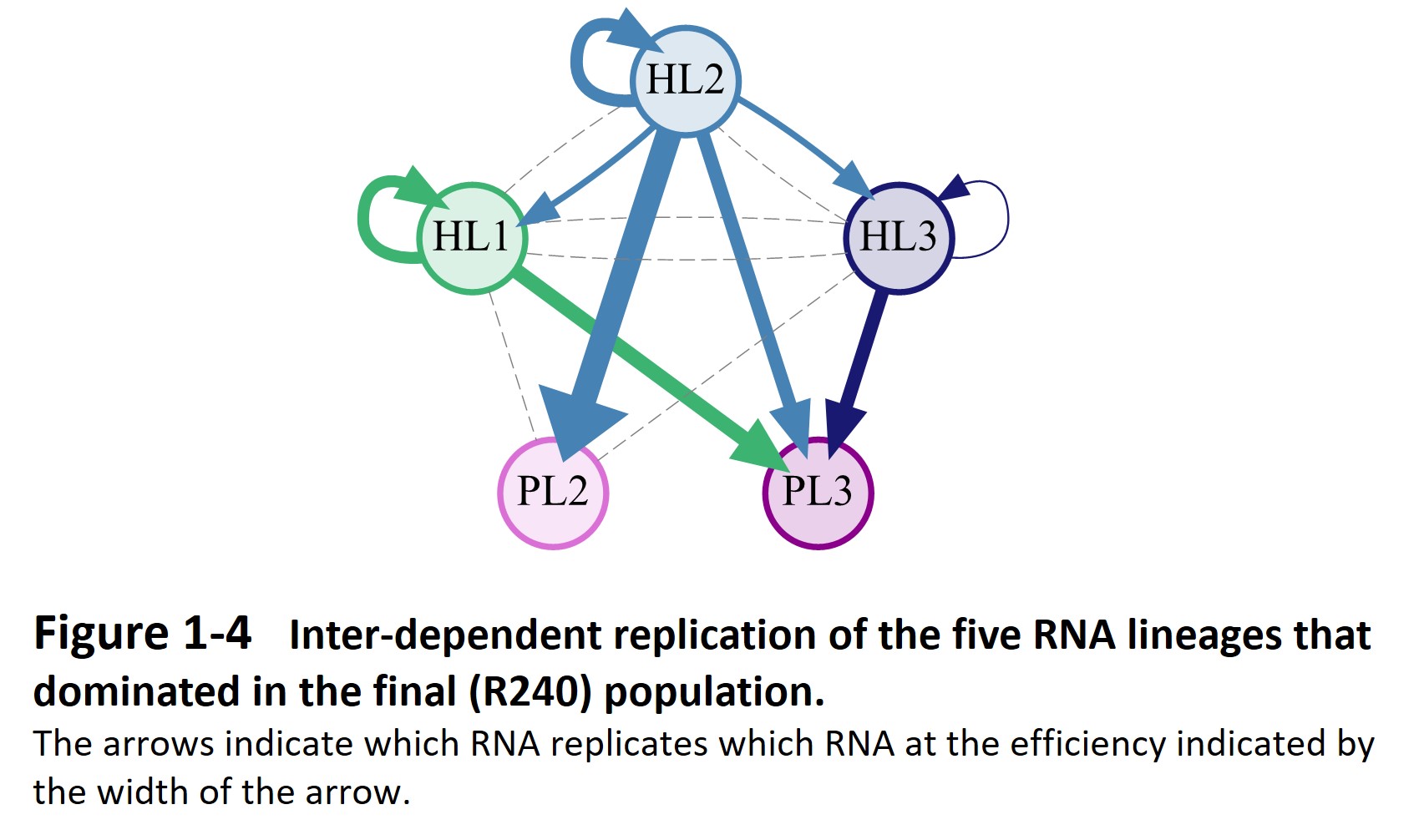
These findings suggest that when a molecular self-replicating system undergoes Darwinian evolution, it can spontaneously diversify and form a complex replication network, which may be the first step toward the emergence of life.
Now, the evolution of RNA is still continuing and producing new phenomena. We hope that the RNA-protein self-replication system can be a useful experimental tool to understand the possible evolutionary process of primitive self-replicators.
Designing and constructing of self-regenerative molecular systems
One of the unique characteristics of living things is recursive self-regeneration. All living organisms or cells are able to reproduce themselves and grow indefinitely. This ability allows us to use microorganisms as a production platform for biomolecules. We are now trying to develop an artificial in vitro system that has this recursive self-regenerative ability for a more controllable and safer platform for biomolecule production. We are working on these projects.
2-1. Development of an artificial DNA self-replication system
Although we have succeeded in developing an RNA genome replication system as the simplest self-replication system, a DNA genome is preferable to encode many genes and replicate with a lower error rate. To date, we have constructed a transcription-translation coupled DNA replication system using phi29 DNA polymerase (Figure 2-1, Sakatani et al. 2015 and 2018). This is the simplest artificial DNA replication system because it requires only two genes for replication, phi29 DNA polymerase and Cre recombinase.
We also found a simpler recursive replication scheme almost by accident. This scheme requires only one gene, the phi29 DNA polymerase gene, and repetitive long linear DNA (Figure 2-2, Okauchi et al. 2020). Normally, DNA polymerase cannot recursively replicate a lnear DNA because the newly synthesized DNA must be shorter than the template DNA according to the "end replication problem". However, if the template DNA consists of repetitive sequences, the product DNA can retain its original size by hybridizing newly synthesized strands at a shifted position. Since this is the simplest DNA replication scheme, we think that this may be a primitive DNA replication scheme.
2-2. In vitro tRNA synthesis
The present artificial cell contains the reconstituted transcription/translation system, which is externally supplied. To achieve complete regeneration of the transcription/translation system, we are trying to synthesize all tRNAs from DNA in the reconstituted system. To date, we have succeeded in simultaneously synthesizing 15 tRNAs from 21 minimal tRNA sets and in coupling tRNA synthesis with the replication of DNA encoding a tRNA gene (Miyachi et al. 2022). We are now trying to synthesize all 21 tRNAs.
2-3. In vitro 20 aaRS regeneration
Another challenge for the complete regeneration of the transcription/translation system is the production of translation proteins, which number 36 in addition to the ribosome. More than half of the translation factors are 20 types of aminoacyl-tRNA synthetases, which attach each amino acid to the corresponding tRNA. To date, we have succeeded in simultaneously expressing 20 aminoacyl-tRNA (aaRS) from the corresponding DNA and coupling synthesis with DNA replication (Fig. 2-3, Hagino et al. 2023). However, the expression efficiency was not sufficient to maintain the initial translation level.
In the subsequent study, we aimed for a sustainable regeneration, which enabled a constant translation activity even after many times of dilution with 20 aaRS-omitting PURE system. We investigated the expression and activities of 20 aaRS when expressed in PURE system, and found that some aaRSs were not sufficiently expressed and the activity was lower than expected. To solve these problem, we furhte optimized the 5'-UTR sequence and adopted another bacterial aaRS with higher activity. After the optimization, we finally suceeded in maintaining the translation activity even after diluting 20 times with 20 aaRS-omitting PURE system, which means that all aaRSs were regenerated to sufficient amounts continuously (Fig. 2-4, Hagino et al. 2025). This is the first step toward the construction of self-regenerative artificial molecular system.
Applications using the constructed systems
3-1. Directed evolution of polymerases and ribosomal RNAs
The artificial cells we have developed are useful as a reactor for directed evolution of RNA and DNA polymerases, as well as rRNA (Sakatani et al. 2019 and Murase et al. 2018). In contrast to conventional evolutionary engineering using microorganisms, our molecular systems allow complete in vitro evolution, which is particularly useful for obtaining mutant enzymes that are toxic to the host cell and also use unnatural cell-impermeable substrates.
References
- Hagino, K., Masuda, K., Shimizu, Y., Ichihashi N.*
Sustainable regeneration of 20 aminoacyl-tRNA synthetases in a reconstituted system toward self-synthesizing artificial systems.
Science Advances 11, eadt6269 (2025) - Hagino, K., Ichihashi, N.*
In vitro transcription/translation-coupled DNA replication through partial regeneration of 20 aminoacyl-tRNA synthetases
ACS Synthetic Biology, 12, 1252–1263 (2023) Full text
- Kamiura, R., Mizuuchi, R., Ichihashi, N.*
Plausible pathway for a host-parasite molecular replication network to increase its complexity through Darwinian evolution
Plos Computational Biology, 18, e1010709 (2022) PDF
- Miyachi, R., Shimizu, Y., Ichihashi, N.*
Transfer RNA Synthesis-Coupled Translation and DNA Replication in a Reconstituted Transcription/Translation System
ACS Synthetic Biology, 11, 2791–2799 (2022) Full text
- Mizuuchi, R.*, Furubayashi, T., Ichihashi, N.* (*co-corresponding)
Evolutionary transition from a single RNA replicator to a multiple replicator network
Nature Communications, 13, 1460 (2022) PDF
- Furubayashi, T., Ueda, K., Bansho, Y., Motooka, D., Nakamura, S., Mizuuchi, R., Ichihashi, N.*
Emergence and diversification of a host-parasite RNA ecosystem through Darwinian evolution
eLIFE 9:e56038 (2020) PDF
- Okauchi, H., Sakatani,Y., Ohtsuka, K., Ichihashi, N.*
Minimization of elements for isothermal DNA replication by an evolutionary approach
ACS Synthetic Biology 9, 1771-1780 (2020) - Sakatani,Y., Mizuuchi, R., Ichihashi, N.*
In vitro evolution of phi29 DNA polymerases through compartmentalized gene expression and rolling-circle replication
Protein Engineering Design and Selection, 32, 481-487 (2019) - Sakatani, Y., Yomo, T., Ichihashi, N.
Self-replication of circular DNA by a self-encoded DNA polymerase through rolling-circle replication and recombination.
Sci Rep 8, 13089 (2018) PDF
- Murase, Y., Nakanishi, H., Tsuji, G., Sunami, S., Ichihashi, N.
In vitro evolution of unmodified 16S rRNA for simple ribosome reconstitution
ACS Synthetic Biology, 7, 576-583 (2018) PDF
- Bansho, Y., Furubayashi, T., Ichihashi, N.*, Yomo, T.* (*co-corresponding)
Host-parasite oscillation dynamics and evolution in a compartmentalized RNA replication system
Proc Nat Sci USA, 113, 4045-4050 (2016) - Sakatani, Y., Ichihashi, N., Kazuta, Y., Yomo, T.
A transcription and translation-coupled DNA replication system using rolling-circle replication
Scientific Reports 5, 10404. (2015) - Ichihashi, N., Usui, K., Kazuta, Y., Sunami, T., Matuura, T., Yomo, T.
Darwinian evolution in a translation-coupled RNA replication system within a cell-like compartment.
Nature Communications, 4, 1-7, (2013) - Yukawa, K., Mizuuchi, R., *Ichihashi, N.
How prebiotic complexity increases through Darwinian evolution.
Curr Opin Syst Biol, 34, 100456 (2023) PDF
- Furubayashi, T., Ichihashi, N.
How evolution builds up complexity?: In vitro evolution approaches to witness complexification in artificial molecular replication systems
Biophysics and Physicobiology, 19, e190005 (2022)
